kopia lustrzana https://github.com/RobertGawron/IonizationChamber
Merge pull request #239 from RobertGawron/feature/travis_to_github_actions_transition
Feature/travis to GitHub actions transitionpull/240/head
commit
49cbbfd875
|
|
@ -0,0 +1 @@
|
|||
python_lint_repport/
|
||||
Plik binarny nie jest wyświetlany.
|
Po Szerokość: | Wysokość: | Rozmiar: 10 KiB |
Plik binarny nie jest wyświetlany.
|
Po Szerokość: | Wysokość: | Rozmiar: 24 KiB |
|
|
@ -0,0 +1 @@
|
|||
__pycache__/
|
||||
|
|
@ -1,20 +1,24 @@
|
|||
from serial import Serial
|
||||
import mcp3425
|
||||
|
||||
|
||||
class IonizationChamber:
|
||||
def __init__(self, config):
|
||||
self.config = config
|
||||
|
||||
|
||||
def connect(self):
|
||||
self.serialPort = Serial(self.config.myDeviceId, baudrate = self.config.myBaudrate, timeout=None)
|
||||
self.serialPort = Serial(
|
||||
self.config.myDeviceId,
|
||||
baudrate=self.config.myBaudrate,
|
||||
timeout=None)
|
||||
|
||||
self.serialPort.isOpen()
|
||||
self.serialPort.flushInput()
|
||||
|
||||
|
||||
def getMeasurement(self):
|
||||
dataIn = self.serialPort.read(5)
|
||||
(msb, lsb) = (dataIn[2], dataIn[3])
|
||||
deviceMeasurement = mcp3425.convert(msb, lsb, mcp3425.MCP3425_RESOLUTION.R14)
|
||||
return deviceMeasurement
|
||||
(msb, lsb) = (dataIn[2], dataIn[3])
|
||||
deviceMeasurement = mcp3425.convert(
|
||||
msb, lsb, mcp3425.MCP3425_RESOLUTION.R14)
|
||||
|
||||
return deviceMeasurement
|
||||
|
|
|
|||
|
|
@ -1,7 +1,52 @@
|
|||
# Firmware
|
||||
|
||||
* ["Data processing and firmware flashing" node architecture
|
||||
](https://github.com/RobertGawron/IonizationChamber/wiki/%22Data-processing-and-firmware-flashing%22-node-architecture
|
||||
)
|
||||
|
||||
## Setup
|
||||
|
||||
* [Setting up development environment on Linux
|
||||
](https://github.com/RobertGawron/IonizationChamber/wiki/Setting-up-development-environment-on-Linux)
|
||||
|
||||
|
||||
## Architecture
|
||||
|
||||
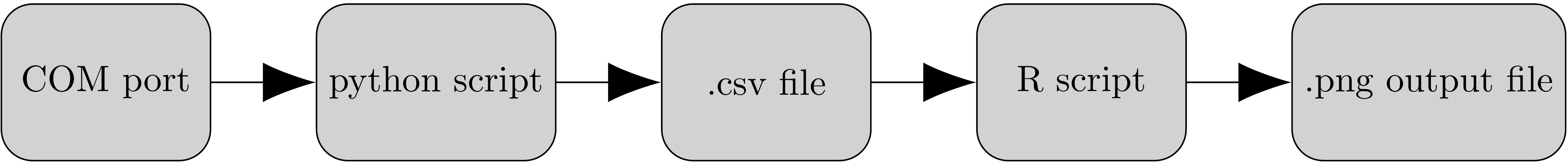
|
||||
|
||||
|
||||
## Collecting measurements
|
||||
|
||||
1. **Edit config.py** to select the correct COM port of Ionization Chamber. Note that **useDMM flag should be set to False**, is experimental and was supposed to be used to check the correlation of Ionization Chamber with other factors (measured by DMM with SCPI support), such factors could be e.g. temperature.
|
||||
|
||||
2. **Run data acquisition script**, it will log Ionization Chamber output on the screen and also it will save it to data.csv for further processing.
|
||||
|
||||
```python main.py```
|
||||
|
||||
3. When all data is logged, terminate ```python main.py``.
|
||||
|
||||
## Plotting signal value in domain time + plotting histogram
|
||||
|
||||
This mode is useful to look on measurement changes over time.
|
||||
|
||||
After collecting data run script to post-process it and generate diagrams:
|
||||
|
||||
```Rscript main.R```
|
||||
|
||||
A new .png image with timestamp in its name will be created in directory where script is.
|
||||
|
||||
Below is example of such generated plot.
|
||||
|
||||
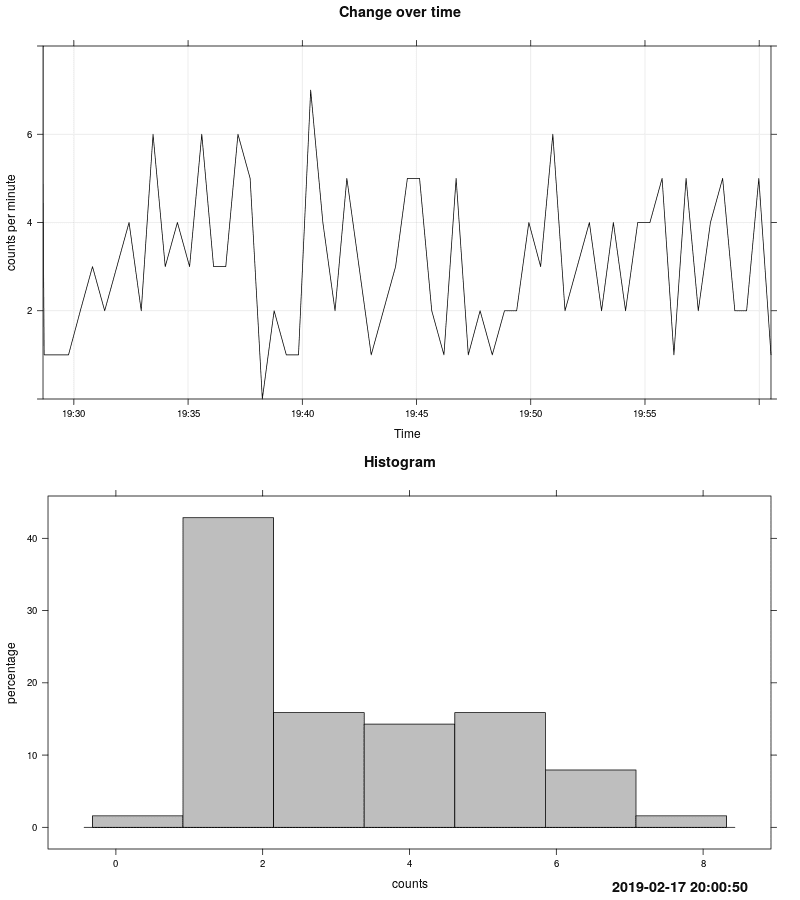
|
||||
|
||||
|
||||
## Plotting values from different measurements [(box plot)](https://en.wikipedia.org/wiki/Box_plot)
|
||||
|
||||
1. Collect data from different samples as different .csv files.
|
||||
2. Edit ```boxplot.R```, to match filenames of .cvs files and labels of measurements.
|
||||
3. Run:
|
||||
|
||||
```Rscript boxplot.R```
|
||||
|
||||
A new .png image with timestamp in its name will be created in directory where script is.
|
||||
|
||||
Below is example of such generated plot.
|
||||
|
||||
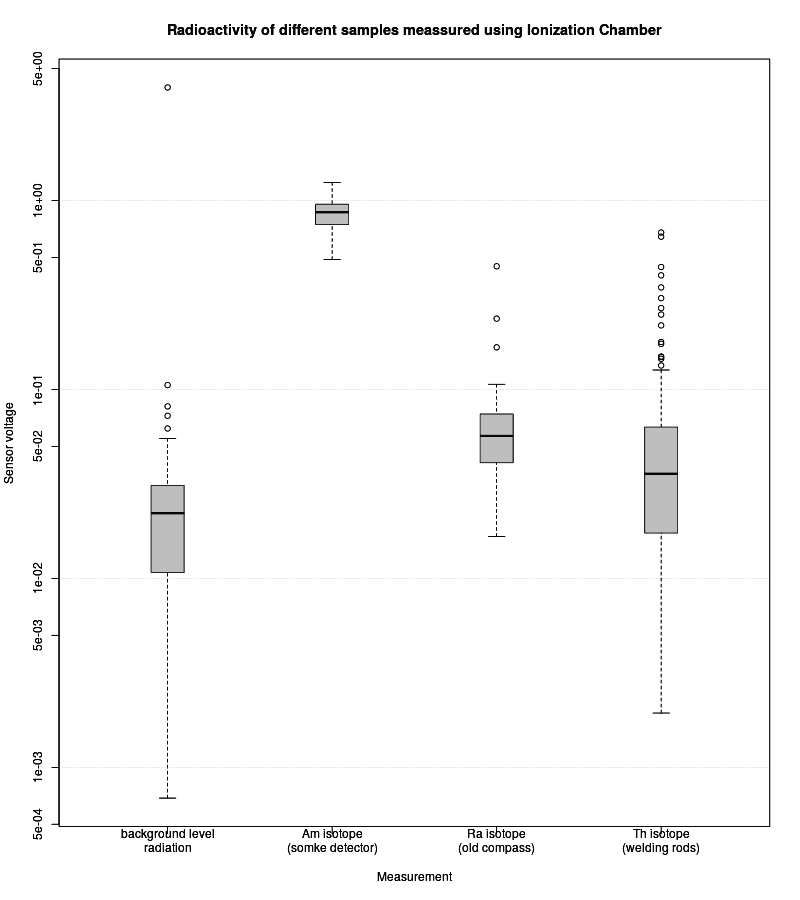
|
||||
|
|
|
|||
|
|
@ -1,10 +1,10 @@
|
|||
inputFileName ="DataNoSample.csv"
|
||||
fileNoSample <- read.delim(inputFileName, , sep=",")
|
||||
|
||||
inputFileName ="DataAmSample1.csv"
|
||||
inputFileName ="DataSample1.csv"
|
||||
fileAmSample1 <- read.delim(inputFileName, , sep=",")
|
||||
|
||||
inputFileName ="DataRaSample2.csv"
|
||||
inputFileName ="DataSample2.csv"
|
||||
fileAmSample2 <- read.delim(inputFileName, , sep=",")
|
||||
|
||||
|
||||
|
|
|
|||
|
|
@ -1,10 +1,9 @@
|
|||
# ionization chamber
|
||||
myDeviceId = '/dev/ttyUSB0'
|
||||
myBaudrate = 9600
|
||||
myBaudrate = 9600
|
||||
|
||||
# dmm
|
||||
idDMM = "USB0::0x2A8D::0x1601::INSTR"
|
||||
testCommand = "READ?"
|
||||
|
||||
|
||||
useDMM = True
|
||||
useDMM = False
|
||||
|
|
|
|||
|
|
@ -1,14 +1,12 @@
|
|||
import usbtmc
|
||||
|
||||
|
||||
class DMM:
|
||||
def __init__(self, config):
|
||||
self.config = config
|
||||
|
||||
|
||||
def connect(self):
|
||||
self.device = usbtmc.Instrument(self.config.idDMM)
|
||||
|
||||
|
||||
def getMeasurement(self):
|
||||
return self.device.ask(self.config.testCommand)
|
||||
|
||||
return self.device.ask(self.config.testCommand)
|
||||
|
|
|
|||
|
|
@ -10,6 +10,6 @@ samples <- read.delim(inputFileName, , sep=",")
|
|||
drawDiagramSingle(samples)
|
||||
|
||||
# TODO move to a separate module
|
||||
library(Hmisc)
|
||||
rcorr(samples$Counter,samples$DMM, type="pearson")
|
||||
#library(Hmisc)
|
||||
#rcorr(samples$Counter,samples$DMM, type="pearson")
|
||||
|
||||
|
|
|
|||
|
|
@ -1,76 +1,70 @@
|
|||
import datetime
|
||||
|
||||
from IonizationChamber import IonizationChamber
|
||||
from IonizationChamber import IonizationChamber
|
||||
from dmm import DMM
|
||||
import config
|
||||
|
||||
|
||||
class IonizationChamberStateMachine:
|
||||
def __init__(self, config):
|
||||
self.config = config
|
||||
self.deviceMeasurement = 0.0
|
||||
self.dmmMeasurement = 0.0
|
||||
|
||||
self.nextState = self.initIonizationChamber
|
||||
|
||||
self.nextState = self.initIonizationChamber
|
||||
|
||||
def tick(self):
|
||||
self.nextState()
|
||||
|
||||
|
||||
def initIonizationChamber(self):
|
||||
self.chamber = IonizationChamber(config)
|
||||
self.chamber.connect()
|
||||
|
||||
|
||||
if config.useDMM:
|
||||
self.nextState = self.initDMM
|
||||
else:
|
||||
self.nextState = self.initOutputFile
|
||||
|
||||
|
||||
def initDMM(self):
|
||||
self.dmm = DMM(config)
|
||||
self.dmm.connect()
|
||||
|
||||
self.nextState = self.initOutputFile
|
||||
|
||||
self.nextState = self.initOutputFile
|
||||
|
||||
def initOutputFile(self):
|
||||
self.logFile = open('data.csv', 'w')
|
||||
self.logFile.write("Time,Counter,DMM\n")
|
||||
|
||||
self.nextState = self.getMeasurementFromIonizationChamber
|
||||
|
||||
self.nextState = self.getMeasurementFromIonizationChamber
|
||||
|
||||
def getMeasurementFromIonizationChamber(self):
|
||||
self.deviceMeasurement = self.chamber.getMeasurement()
|
||||
|
||||
|
||||
if config.useDMM:
|
||||
self.nextState = self.getMeasurementFromDMM
|
||||
else:
|
||||
self.nextState = self.saveMeasurement
|
||||
|
||||
|
||||
def getMeasurementFromDMM(self):
|
||||
self.dmmMeasurement = self.dmm.getMeasurement()
|
||||
|
||||
self.nextState = self.saveMeasurement
|
||||
|
||||
self.nextState = self.saveMeasurement
|
||||
|
||||
def saveMeasurement(self):
|
||||
now = datetime.datetime.now()
|
||||
self.logFile.write("{0},{1},{2}\n".format(now, self.deviceMeasurement, self.dmmMeasurement))
|
||||
self.logFile.flush()
|
||||
|
||||
self.nextState = self.showMeasurementToUser
|
||||
self.logFile.write("{0},{1},{2}\n".format(
|
||||
now, self.deviceMeasurement, self.dmmMeasurement))
|
||||
|
||||
self.logFile.flush()
|
||||
|
||||
self.nextState = self.showMeasurementToUser
|
||||
|
||||
def showMeasurementToUser(self):
|
||||
print("{0}, {1}".format(self.deviceMeasurement, self.dmmMeasurement))
|
||||
|
||||
|
||||
self.nextState = self.getMeasurementFromIonizationChamber
|
||||
|
||||
|
||||
if __name__=="__main__":
|
||||
if __name__ == "__main__":
|
||||
machine = IonizationChamberStateMachine(config)
|
||||
|
||||
while True:
|
||||
|
|
|
|||
|
|
@ -1,24 +1,27 @@
|
|||
# based on https://ww1.microchip.com/downloads/en/DeviceDoc/22072b.pdf
|
||||
# assumed that gain = 1
|
||||
|
||||
"""
|
||||
based on https://ww1.microchip.com/downloads/en/DeviceDoc/22072b.pdf
|
||||
assumed that gain = 1
|
||||
"""
|
||||
from enum import Enum
|
||||
|
||||
|
||||
class MCP3425_RESOLUTION(Enum):
|
||||
R12 = 1
|
||||
R13 = 2
|
||||
R14 = 3
|
||||
|
||||
def convert(upperByte, lowerByte, resolution):
|
||||
digitalToAnalog = lambda value, lsb, pga: (value * (lsb / pga))
|
||||
|
||||
digitalOutput = (upperByte << 8) | lowerByte;
|
||||
def convert(upperByte, lowerByte, resolution):
|
||||
def digitalToAnalog(value, lsb, pga):
|
||||
return (value * (lsb / pga))
|
||||
|
||||
digitalOutput = (upperByte << 8) | lowerByte
|
||||
|
||||
if resolution == MCP3425_RESOLUTION.R12:
|
||||
return digitalToAnalog(digitalOutput, (1 * 0.01), 1)
|
||||
|
||||
|
||||
if resolution == MCP3425_RESOLUTION.R13:
|
||||
return digitalToAnalog(digitalOutput, (250 * 0.0000001), 1)
|
||||
|
||||
|
||||
if resolution == MCP3425_RESOLUTION.R14:
|
||||
return digitalToAnalog(digitalOutput, (62.5 * 0.000001), 1)
|
||||
|
||||
|
|
|
|||
Ładowanie…
Reference in New Issue